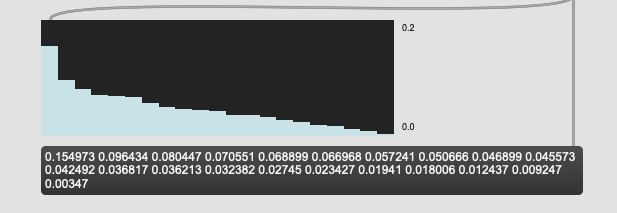
Ok, it turns out I wasn’t normalizing the dataset before the PCA-ing, so in doing that I get the same weights as you now (well, diff float resolution):
0.154973 0.096434 0.080447 0.070551 0.068899 0.066968 0.057241 0.050666 0.046899 0.045573 0.042492 0.036817 0.036213 0.032382 0.02745 0.023427 0.01941 0.018006 0.012437 0.009247 0.00347
I still get shitty results after that step though.
If i multiply those weights by each element of the transposed (rotated) bases I now get this:
1, 0.085675 0.008393 0.001326 0.008187 0.007971 0.008628 0.013086 0.004254 0.002104 0.000748 0.004894 0.00036 0.001792 0.000635 0.000365 0.003239 0.002782 0.000216 0.008884 0.000597 0.000361;
2, 0.007919 0.007471 0.006706 0.040497 0.016871 0.009139 0.006336 0.007237 0.006255 0.017774 0.015751 0.007725 0.003578 0.010975 0.000198 0.003128 0.000263 0.003946 0.000041 0.000033 0.00014;
3, 0.052043 0.001921 0.000773 0.021265 0.008598 0.002079 0.005835 0.000392 0.006376 0.001308 0.004743 0.008552 0.00722 0.002093 0.001648 0.015659 0.004895 0.00475 0.002496 0.001147 0.000019;
4, 0.031974 0.022968 0.004003 0.001664 0.01861 0.004415 0.029216 0.013642 0.012323 0.001535 0.001403 0.018025 0.00695 0.001735 0.003461 0.00596 0.001939 0.000042 0.00262 0.000382 0.000004;
5, 0.045223 0.002707 0.007318 0.026383 0.005528 0.016972 0.019099 0.004125 0.007298 0.000378 0.017384 0.01283 0.002749 0.006888 0.001546 0.003838 0.002201 0.007323 0.00027 0.000619 0.000009;
6, 0.002331 0.038066 0.001567 0.001518 0.025555 0.016394 0.006657 0.015385 0.005177 0.023422 0.008098 0.00586 0.000801 0.007785 0.002735 0.005511 0.001809 0.004711 0.000332 0.000092 0.00007;
7, 0.003937 0.011633 0.019493 0.017428 0.001973 0.00893 0.017583 0.006289 0.002516 0.00732 0.008437 0.006867 0.016423 0.001173 0.010752 0.000511 0.003452 0.007802 0.002297 0.001273 0.000077;
8, 0.00387 0.045505 0.011949 0.013447 0.016632 0.007788 0.001185 0.002647 0.003663 0.005656 0.015184 0.009292 0.009916 0.015145 0.004242 0.003061 0.004089 0.002635 0.000627 0.001464 0.00017;
9, 0.014788 0.035159 0.034758 0.001285 0.0118 0.001598 0.004427 0.015564 0.018955 0.003427 0.004699 0.013617 0.00332 0.002438 0.002516 0.001481 0.004133 0.004965 0.001804 0.00204 0.000249;
10, 0.008042 0.020211 0.038046 0.005854 0.00503 0.00662 0.000684 0.011239 0.002538 0.022968 0.005309 0.00633 0.00962 0.007491 0.005555 0.005341 0.00237 0.002592 0.000283 0.002759 0.000412;
11, 0.002205 0.038956 0.016021 0.00144 0.025907 0.014119 0.004218 0.00548 0.002116 0.004642 0.001317 0.009105 0.014186 0.000763 0.009008 0.001934 0.006157 0.002039 0.000826 0.003047 0.000539;
12, 0.006574 0.008713 0.038988 0.010971 0.008535 0.013974 0.009623 0.017562 0.001279 0.007431 0.013808 0.004971 0.008979 0.005301 0.000321 0.005214 0.002369 0.003255 0.000071 0.00364 0.000621;
13, 0.006309 0.001324 0.011464 0.003034 0.018929 0.033917 0.004756 0.016611 0.012228 0.011017 0.007867 0.000306 0.000959 0.000293 0.011525 0.003128 0.001633 0.000002 0.000483 0.002769 0.000954;
14, 0.00479 0.01835 0.016558 0.000861 0.014925 0.023515 0.018183 0.001237 0.009726 0.014072 0.00404 0.004132 0.00028 0.012425 0.005908 0.000857 0.003898 0.003538 0.00091 0.002799 0.001191;
15, 0.010411 0.000802 0.016985 0.009138 0.006162 0.019901 0.004118 0.003413 0.01212 0.004181 0.002088 0.000008 0.004302 0.013141 0.003365 0.005 0.009029 0.003545 0.000867 0.000049 0.001738;
16, 0.003376 0.029955 0.005552 0.001059 0.007522 0.025932 0.017323 0.000428 0.013371 0.002039 0.000239 0.000393 0.012889 0.006674 0.012893 0.000401 0.001335 0.000208 0.000703 0.001166 0.001331;
17, 0.005899 0.010194 0.005976 0.018071 0.000017 0.00917 0.017292 0.013394 0.019993 0.009638 0.002888 0.006819 0.000199 0.003858 0.000498 0.001016 0.004162 0.001705 0.000375 0.003888 0.001634;
18, 0.002303 0.016692 0.017512 0.004885 0.02297 0.015379 0.002892 0.026624 0.014126 0.003872 0.002361 0.002061 0.000263 0.005358 0.001792 0.003526 0.007392 0.002612 0.000589 0.002668 0.000884;
19, 0.012102 0.010307 0.016405 0.025596 0.025135 0.001103 0.014636 0.00953 0.002035 0.007411 0.002585 0.011326 0.014214 0.002708 0.008104 0.000872 0.006245 0.000522 0.000019 0.002345 0.000603;
20, 0.080493 0.001526 0.001882 0.004281 0.004025 0.00079 0.002943 0.002856 0.007404 0.00084 0.01268 0.000911 0.003213 0.00646 0.005477 0.008516 0.002729 0.004724 0.006727 0.000245 0.000003;
21, 0.060185 0.00374 0.007563 0.018859 0.015962 0.008397 0.011263 0.005141 0.016239 0.001194 0.018245 0.005911 0.00593 0.005143 0.005687 0.003008 0.00432 0.006233 0.002177 0.000752 0.000049;
22, 0.085675 0.008393 0.001326 0.008187 0.007971 0.008628 0.013086 0.004254 0.002104 0.000748 0.004894 0.00036 0.001792 0.000635 0.000365 0.003239 0.002782 0.000216 0.008884 0.000597 0.000361;
23, 0.007919 0.007471 0.006706 0.040497 0.016871 0.009139 0.006336 0.007237 0.006255 0.017774 0.015751 0.007725 0.003578 0.010975 0.000198 0.003128 0.000263 0.003946 0.000041 0.000033 0.00014;
24, 0.052043 0.001921 0.000773 0.021265 0.008598 0.002079 0.005835 0.000392 0.006376 0.001308 0.004743 0.008552 0.00722 0.002093 0.001648 0.015659 0.004895 0.00475 0.002496 0.001147 0.000019;
25, 0.031974 0.022968 0.004003 0.001664 0.01861 0.004415 0.029216 0.013642 0.012323 0.001535 0.001403 0.018025 0.00695 0.001735 0.003461 0.00596 0.001939 0.000042 0.00262 0.000382 0.000004;
26, 0.045223 0.002707 0.007318 0.026383 0.005528 0.016972 0.019099 0.004125 0.007298 0.000378 0.017384 0.01283 0.002749 0.006888 0.001546 0.003838 0.002201 0.007323 0.00027 0.000619 0.000009;
27, 0.002331 0.038066 0.001567 0.001518 0.025555 0.016394 0.006657 0.015385 0.005177 0.023422 0.008098 0.00586 0.000801 0.007785 0.002735 0.005511 0.001809 0.004711 0.000332 0.000092 0.00007;
28, 0.003937 0.011633 0.019493 0.017428 0.001973 0.00893 0.017583 0.006289 0.002516 0.00732 0.008437 0.006867 0.016423 0.001173 0.010752 0.000511 0.003452 0.007802 0.002297 0.001273 0.000077;
29, 0.00387 0.045505 0.011949 0.013447 0.016632 0.007788 0.001185 0.002647 0.003663 0.005656 0.015184 0.009292 0.009916 0.015145 0.004242 0.003061 0.004089 0.002635 0.000627 0.001464 0.00017;
30, 0.014788 0.035159 0.034758 0.001285 0.0118 0.001598 0.004427 0.015564 0.018955 0.003427 0.004699 0.013617 0.00332 0.002438 0.002516 0.001481 0.004133 0.004965 0.001804 0.00204 0.000249;
31, 0.008042 0.020211 0.038046 0.005854 0.00503 0.00662 0.000684 0.011239 0.002538 0.022968 0.005309 0.00633 0.00962 0.007491 0.005555 0.005341 0.00237 0.002592 0.000283 0.002759 0.000412;
32, 0.002205 0.038956 0.016021 0.00144 0.025907 0.014119 0.004218 0.00548 0.002116 0.004642 0.001317 0.009105 0.014186 0.000763 0.009008 0.001934 0.006157 0.002039 0.000826 0.003047 0.000539;
33, 0.006574 0.008713 0.038988 0.010971 0.008535 0.013974 0.009623 0.017562 0.001279 0.007431 0.013808 0.004971 0.008979 0.005301 0.000321 0.005214 0.002369 0.003255 0.000071 0.00364 0.000621;
34, 0.006309 0.001324 0.011464 0.003034 0.018929 0.033917 0.004756 0.016611 0.012228 0.011017 0.007867 0.000306 0.000959 0.000293 0.011525 0.003128 0.001633 0.000002 0.000483 0.002769 0.000954;
35, 0.00479 0.01835 0.016558 0.000861 0.014925 0.023515 0.018183 0.001237 0.009726 0.014072 0.00404 0.004132 0.00028 0.012425 0.005908 0.000857 0.003898 0.003538 0.00091 0.002799 0.001191;
36, 0.010411 0.000802 0.016985 0.009138 0.006162 0.019901 0.004118 0.003413 0.01212 0.004181 0.002088 0.000008 0.004302 0.013141 0.003365 0.005 0.009029 0.003545 0.000867 0.000049 0.001738;
37, 0.003376 0.029955 0.005552 0.001059 0.007522 0.025932 0.017323 0.000428 0.013371 0.002039 0.000239 0.000393 0.012889 0.006674 0.012893 0.000401 0.001335 0.000208 0.000703 0.001166 0.001331;
38, 0.005899 0.010194 0.005976 0.018071 0.000017 0.00917 0.017292 0.013394 0.019993 0.009638 0.002888 0.006819 0.000199 0.003858 0.000498 0.001016 0.004162 0.001705 0.000375 0.003888 0.001634;
39, 0.002303 0.016692 0.017512 0.004885 0.02297 0.015379 0.002892 0.026624 0.014126 0.003872 0.002361 0.002061 0.000263 0.005358 0.001792 0.003526 0.007392 0.002612 0.000589 0.002668 0.000884;
40, 0.012102 0.010307 0.016405 0.025596 0.025135 0.001103 0.014636 0.00953 0.002035 0.007411 0.002585 0.011326 0.014214 0.002708 0.008104 0.000872 0.006245 0.000522 0.000019 0.002345 0.000603;
41, 0.080493 0.001526 0.001882 0.004281 0.004025 0.00079 0.002943 0.002856 0.007404 0.00084 0.01268 0.000911 0.003213 0.00646 0.005477 0.008516 0.002729 0.004724 0.006727 0.000245 0.000003;
42, 0.060185 0.00374 0.007563 0.018859 0.015962 0.008397 0.011263 0.005141 0.016239 0.001194 0.018245 0.005911 0.00593 0.005143 0.005687 0.003008 0.00432 0.006233 0.002177 0.000752 0.000049;
Which now gives me the same same, but different, but still same final results of:
ImportantFeatures: MFCC1
ImportantFeatures: MFCC2
ImportantFeatures: MFCC3
ImportantFeatures: MFCC5
ImportantFeatures: MFCC6
ImportantFeatures: MFCC4
ImportantFeatures: MFCC7
ImportantFeatures: MFCC8
ImportantFeatures: MFCC9
ImportantFeatures: MFCC11
It kind of feels like theres some other rotation I’m missing later in the process.
If I take the bases and sum them together willy nilly I get this:
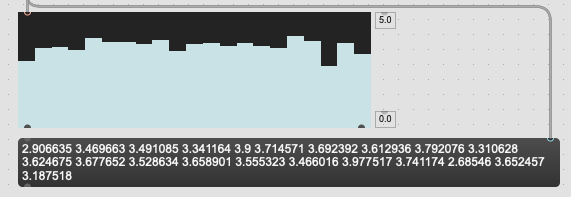
Kind of flat-ish.
And the results of my (and your) weights returns this:
So the aggregate of multiplying those together (or rather, multiplying each base by the weighting) can only really return me this:
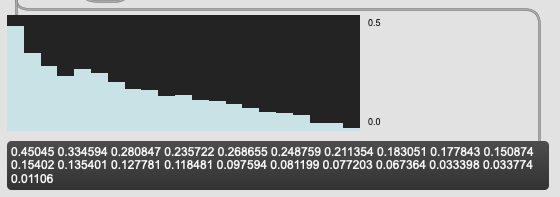
On a hunch I’ve tried this on a couple other datasets and I’m getting results that look like bugs. Will make a new thread about it.