finding things in stuff
In the last flesh flucoma plenary I presented some work that I did combining FluCoMa, Python and composition. The title of that talk “Finding things in stuff” has permeated my interests since then, and I sought to develop a set of tools that would last forever for me, drawing together all of the techniques and implementations I found useful into one streamlined workflow. Last night I uploaded the first stable version of that software ‘ftis’ which I have resigned will likely just be interesting and useful for me  but nonetheless is now public and relatively usable by anyone interested. Publishing it to the public repository of
but nonetheless is now public and relatively usable by anyone interested. Publishing it to the public repository of pip installable modules is a way of closing off that project for a while now so I thought I would share it here.
example
In light of the recent geek-out session on Thursday afternoon I thought I would use an example of what ftis can do and how it pulls together various technologies to provide a fast, and convenient way of scripting out the data analysis portion of what many people seem to be doing right now (I’m thinking of you @spluta and your frustrations). In this case, I’m going to produce a flucoma compatible dataset from the statistics of chroma analysis.
# import analysers
from ftis.analyser.descriptor import Chroma
from ftis.analyser.audio import CollapseAudio
from ftis.analyser.stats import Stats
# import scaffolding
from ftis.corpus import Corpus
from ftis.process import FTISProcess
from ftis.common.io import write_json
from pathlib import Path
# import the necessary flucoma materials
from flucoma import dataset
src = Corpus("~/corpus-folder/corpus1")
out = "~/corpus-folder/chroma-dataset"
process = FTISProcess(source=src, folder=out)
stats = Stats(numderivs=2, spec=["stddev", "mean"]) # use a non-anonymous class
process.add(
CollapseAudio(),
Chroma(fmin=40),
stats
)
if __name__ == "__main__":
process.run()
# Now that ftis has completed lets pack the data into a fluid dataset
dataset = dataset.pack(stats.output) # use the pack function to marshall it to the right format
dataset_path = Path(out) / "dataset.json" # create an output path
write_json(dataset_path.expanduser(), dataset) # write to disk
This script would analyse every audio file inside the src folder and produce a dataset containing statistics on the Chroma analysis. This uses librosa for the Chroma analysis,flucoma for the slicing, pydub for the exploding/collapsing of audio and scipy for the statistics. All of this is multi-threaded and pretty fast. For example, running the above script on a corpus of 1000 items completes for me in about 3 minutes. ftis also implements both high level and low level caching, so individual analyses are stored in between runs. This means you can modify the original code or add additional processes after the analysis, re-run the script and you don’t have to wait on analysis that was already done just before. It also stores all steps of the analysis as simple .json files and if they exist it uses that to immediately load all the analysis as a chunk.
For example, the output of running that script looks like this in the file system:
That dataset.json file can be read into SC, pd, Max or wherever you prefer and all of the analysis files in .json format can be read back into new ftis processes.
I’ve also incorporated more novel dimension reduction algorithms, clustering and even a web server that boots to visually browse your data once made (very very alpha still though). The main interface however is currently terminal based and looks a little like this:
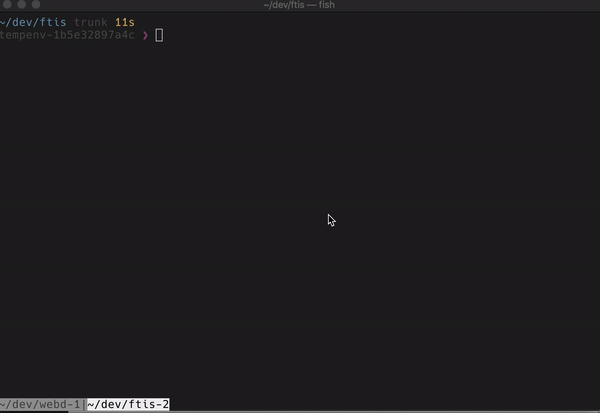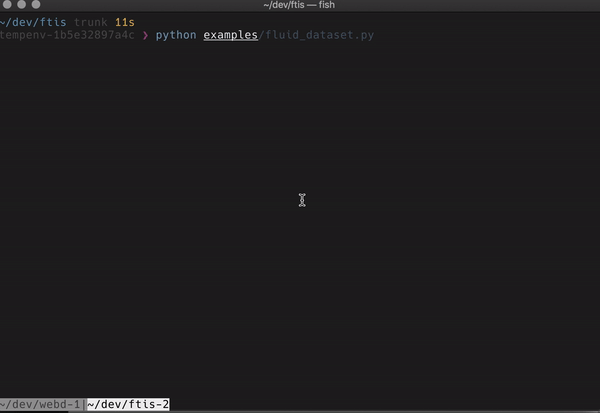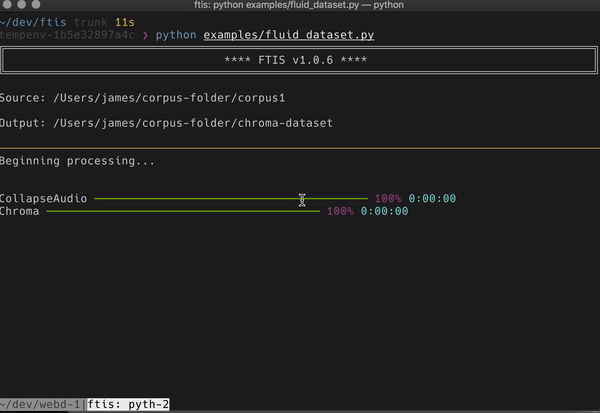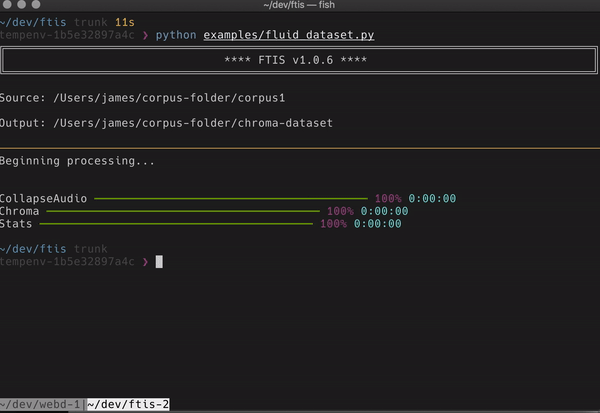
If anyone with some python chops wants to try it out, that would be awesome. I found with ReaCoMa that when other people used it, it became much more robust and useful for both myself and a wider community. My hope is that although ftis is very specialised right now that it could reach a wider audience of people who want to work in a robust way on big datasets that doesn’t break their existing workflow and comfortable ways of musicking.
If you do want to use it, its requires you to run pip install ftis and then look at the examples directory in the source code. I’d be more than up for working with anyone who had a specific goal in mind that they think ftis might help you achieve.