Hello folks, thanks for the amazing work on this toolkit, finally I had some time to play with it and it’s brilliant!
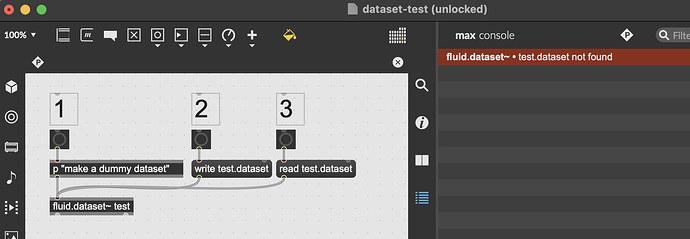
I have a relatively simple mosaicing patch (90% still the original helpfile) for MFCC analysis of a bunch of slices, and I want to save the dataset so that I don’t have to run the analysis every time I open the patch, so I was trying to write-read the data like I do with all buffers there, but the read part looks broken:
The file is created, but the I get an error that file doesn’t exist, even though it’s in the same folder as the patch. (see screenshots attached)
While double-checking everything alongsite writing this post, I have noticed that the “read” message on its own only allows me to select .json or .maxpat files in the select file dialog, and indeed when writing and reading a .json file, everything works as expected. I found this quite confusing because I can read and write buffers~ from/to files of any extensions (and like to use .buffer for data buffers), so it took me quite a while to figure this out.
I’m using OS X Ventura - 13.2.1 (22D68), flucoma 1.0.6, Max 5.8.2.