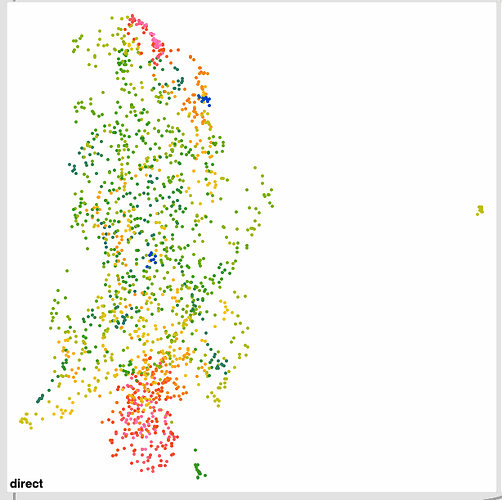
In this weeks geekout we got to talking about the thing I built in this thread and @tremblap mentioned that he wasn’t seeing such good clustering with the MFCC space of 76D (mean/std/min/max) → 4D reduction I was applying. I didn’t see a massive correlation either, but I chalked it up to it being a 76D->4D->3D reduction that was making it shitty.
After some chat (with helpful input from @jamesbradbury and @tedmoore, as well as @tutschku) I decided to revisit that part of the patch, but by figuring out what UMAP settings to use by visualizing 2D/3D reductions of the 76D space, with the idea of then applying these same UMAP settings to the 4D reduction.
There was also some discussion about standardizing before UMAP or feeding the output of MFCCs directly into UMAP.
Here are my tests/results so far (with a video below).
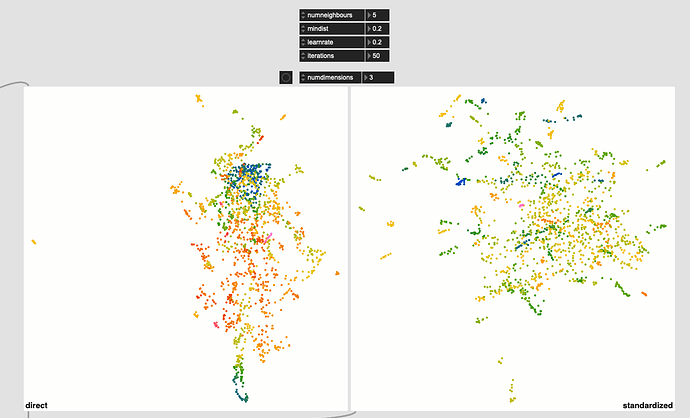
So using the default UMAP settings I was using before (pulled from the helpfile) I get this:
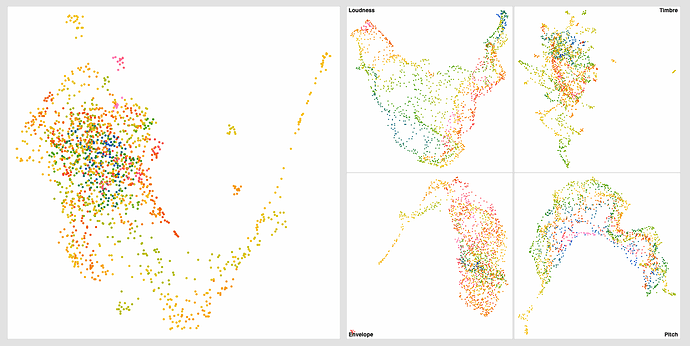
The standardized one (on the right) has more of that stringy/stretched thing I’ve been seeing, which I thought was a “double UMAP” artefact.
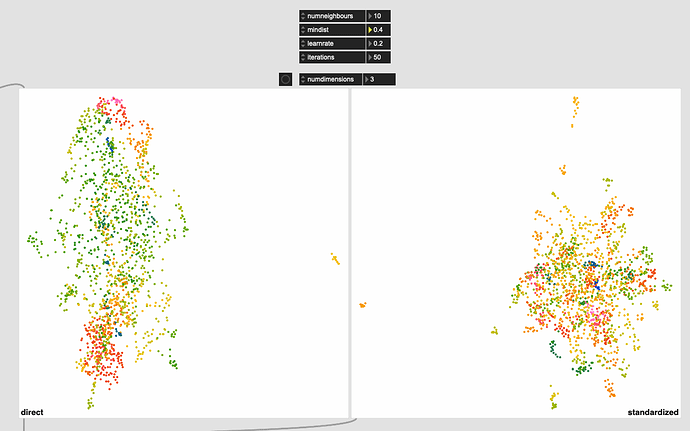
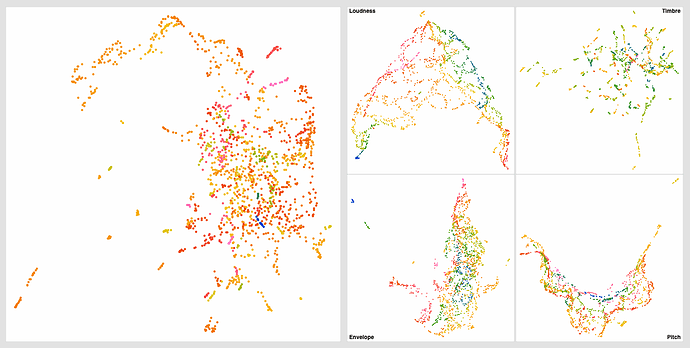
Even with this I get much better ‘clustering’ without the pre-standardization, but in massaging the settings a bit more I get this:
Obviously some weird outliers there, but I suspect that’s due to the dataset being normalized completely for visualization. If I understand fluid.robustscale~ correctly, the main clump on the left will occupy the main fluid.kdtree~ space, with these outliers existing, but being waaaaaay out there.
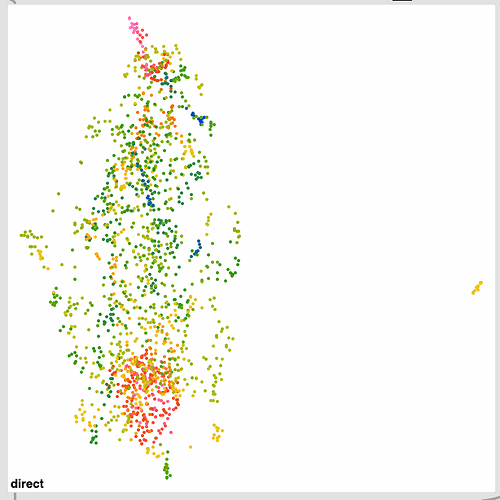
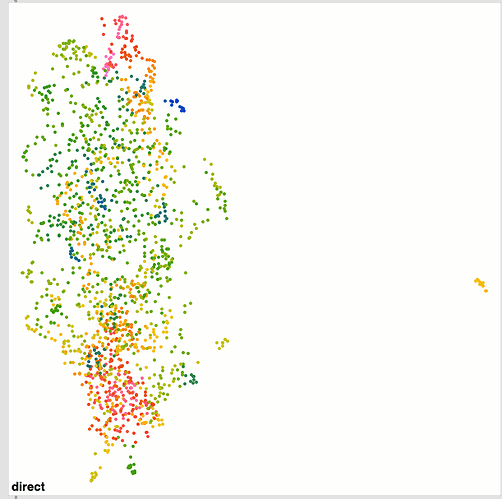
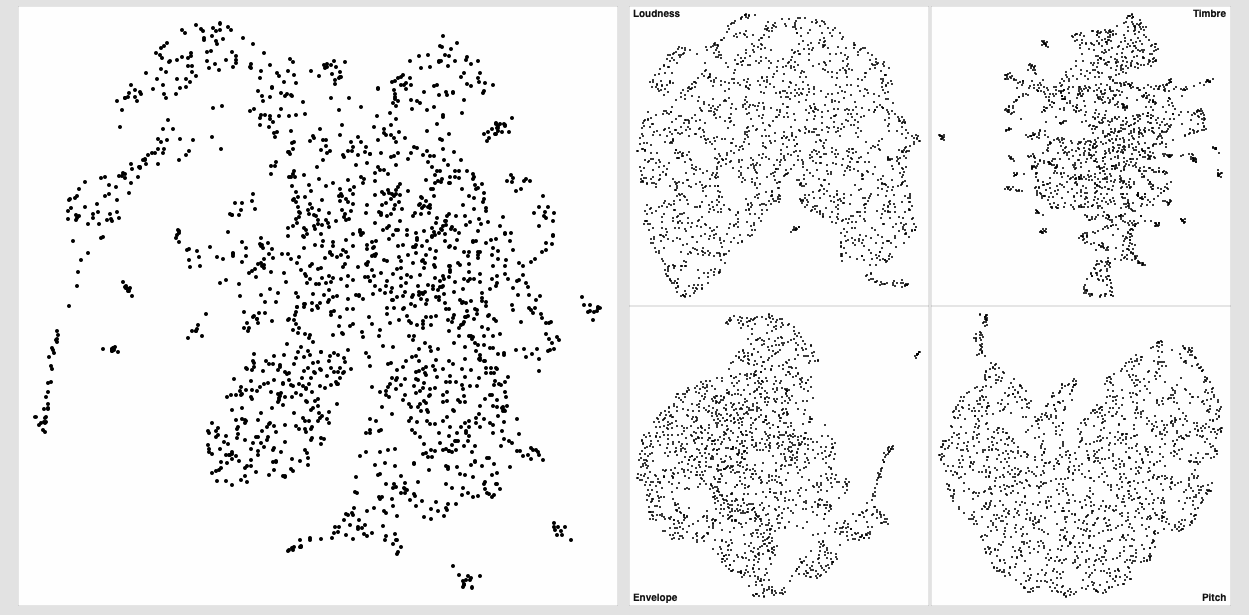
I also ran the same thing over and over again to see how much it converges and it’s pretty good. Wiggles around a bit, but nothing super dramatic. I’m not entirely sure how much movement is “too much” in that sense.
Here’s a bunch of examples of the same dataset being refit:
Here’s a quick video:
////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////
So in addition to just sharing the results of this (which have been great), I wanted to talk about visualizing reductions (particularly when your target is >3), and how generalizable UMAP settings can be.
With regards to the first one, I hadn’t really considered tweaking settings in 3D and then extrapolating that back up to my target dimensionality (4D in this case). I was just relying on 76D → 4D → 3D, and expecting that process to go badly along the way.
As far as the latter, as you can hear from the video, these sounds are roughly in the same ballpark of sounds. Metalic-ish percussive sounds, and all analyzing with tiny windows. So perhaps the UMAP settings above work well for this particular corpus, but may be problematic for other datasets. This will be less cumbersome if some kind of dynamic “analysis pipeline” is implemented where each individual corpus can be tailered and massaged around, and a corresponding realtime/matching analysis pipeline can be reconstructed around those settings, but I’d like to figure out some settings that “work” (well enough) across the board.