Hello,
I am training a mlp regressor to control parameters based on fft frames. Because of the amount of training material and values per point, training takes to long to be feasable to do locally (> 20h). I built the prototype in Max, now I’m wondering if there would be any improvements regarding the training duration if I used SuperCollider (or SuperNova)? The main bottleneck seems to be that in Max the training is only happening on one cpu core, would that be the same in SuperCollider?
Another option would be to train the model in python using sklearn, since there is already a script to convert the sklearn model to the flucoma json format (FluCoMa-stuff/sklearn_mlp_to_fluid_mlp.py at master · tedmoore/FluCoMa-stuff · GitHub). Is there also already a script somewhere to convert the flucoma dataset to matrices as training inputs for sklearn?
I don’t have one, but I reckon @tedmoore has one. Let us know if you manage to make a simple workflow, and why not share it here?
1 Like
There would be no improvements and flucoma doesn’t work with supernova.
I think what I’ve done in the past is export a CSV from supercollider and then consume the csv into python.
If you find a workflow that works for you please do share it here!
1 Like
I might have done a script long ago loading the json file (which is what a dataset is) and parse it in python, but I cannot find it… very likely I pinched it from @weefuzzy or @jamesbradbury or @groma
as I am a python noob
1 Like
Thank you for the replys! I’ll try to put together a workflow, and once I have something that works, I’ll definitely share it.
The link does not seem to be working. 
I think our json export feature is most probably the simplest way to parse a fluiddataset in python. I even think this is why we did it 
1 Like
Oops. Here’s the file.
ArrayToCSV.sc.zip (1.1 KB)
1 Like
I’ve put together a small python script that converts the flucoma datasets to numpy, trains a MLP in scikit-learn and uses the python function from @tedmoore to convert the resulting model back to flucoma:
3 Likes
Nice. I haven’t tested it, but looks cool.
It looks like you’re using the default number of hidden layers (1 layer with 100 neurons) which might be too many! It would be great to modify the script to put some of these arguments into argparse (hiddenlayers, learning rate, activation fuction, etc.) so the interface is more similar to FluCoMa’s (so that FluCoMa users will recognize and use it similarly!)
T
1 Like
Ah yes, that makes sense, I will do that!
2 Likes
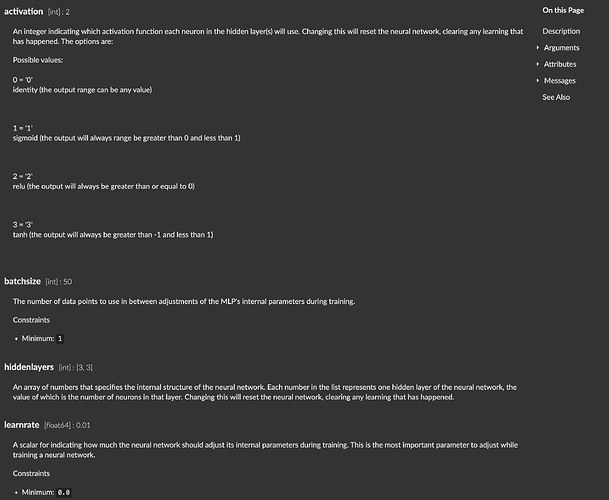
@tedmoore the references do not seem to mention the default values for most of the paramaters. Is there any place to look them up?
sklearn references or flucoma references?
The defaults for the flucoma objects can be found in the Max reference file:
(this isn’t all of them, just a screengrab)
1 Like
I added the command line arguments 
1 Like