Hi,
Thanks a lot for the update. I understand that quite a few people have been using the UMAP and have not found any practical problems with it… So, I may be using inappropriately the transform. Let me first explain the use case I have in mind.
I have a database of piano recordings with about 10 playing techniques which lead to quite different timbres. I characterize the timbres by computing mfcc coefficients and some spectral features in between two successive onsets. The statistical analysis of these descriptors leads to more than 100 values per interval and I would like to represent the timbre as a point in a 3D space. So, I use the UMAP tool to create a mapping between the 100+ descriptors extracted from the audio to the 3D space. This results in 3D points cloud and I can see how the 10 different timbres are more or less clustered together in the 3D space:

3D representation of the timbre. Each color represent a specific timbre resulting from a playing technique
Once this mapping is defined, I would like to use it to characterize the sound of a piano in the context of a live event. As I would like to use the same mapping as the one trained on the database, I was thinking to use the transformpoint message to see whether the current timbre of the piano was close to the representation of the timbre extracted for the database. But doing this I also noticed that event if was using as live piano the audio that was actually used during the training, the mapping was somewhat inaccurate.
So I started to look more closely on a simple case. In the following patch, I am creating a toy dataset of 20 points with 3 values and I use UMAP with fittransform message to project them in a 2D space TestUmap.maxpat (41.6 KB). Once the mapping is defined, I map again all the individual points of the dataset with the transformpoint message. Here is a comparison between the mapping with fittransform and the one with transformpoint. As you can see they are related but quite different
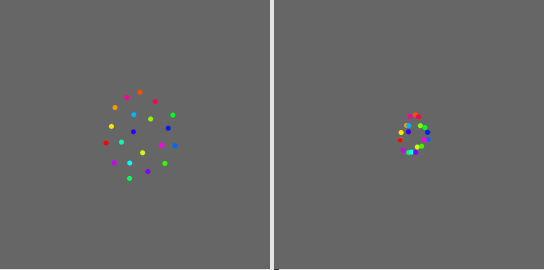
Left: fittransform mapping, right: transformpoint mapping
I understand that there is a stochastic aspect in the transform. Each time I apply the fittransform I get a slightly different result but the relative positions of the points are the same. Here are three additional examples.
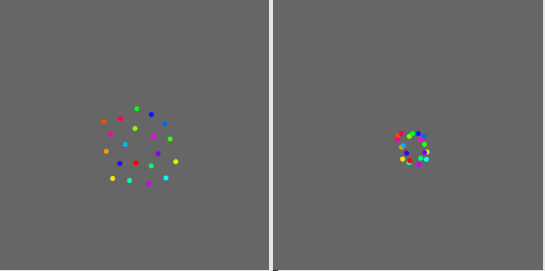
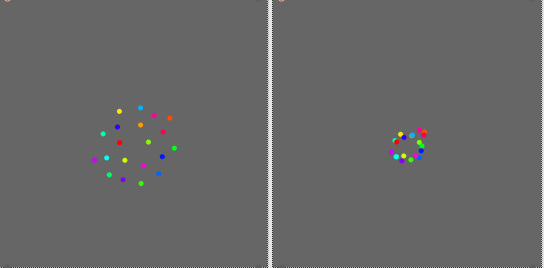

Now what strikes me most in this experiment is that the result of the transformpoint is not simply a small random fluctuation around the fittransform results. There seems to have a constant bias in the shape. In particular the range of values seems to be always smaller with the transformpoint results.
So, I am somewhat confused. I am doing something wrong with the transform or is it really how the transform is supposed to work?
As always, thanks in advance for your feedback and comments.