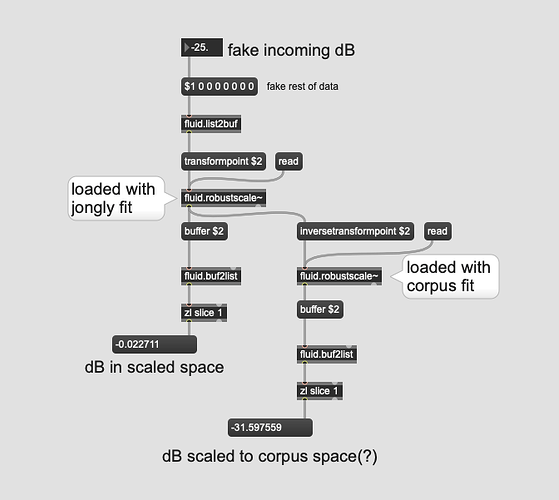
Ok, had some time today to analyze/scale all the bits. So to ask more concretely.
Here is the fluid.robustscale~ fit for the corpus I’m working/testing with:
{
"cols": 8,
"data_high": [
-20.102773666381836,
0.284235775470734,
74.88372039794922,
0.619443297386169,
-28.400463104248047,
0.693569839000702,
73.83578491210938,
0.819302082061768
],
"data_low": [
-41.67296600341797,
-0.263175576925278,
61.05841827392578,
-0.63897043466568,
-67.24171447753906,
-0.872315585613251,
60.706329345703125,
0.185682728886604
],
"high": 75.0,
"low": 25.0,
"median": [
-31.107669830322266,
-0.001829719520174,
68.79005432128906,
-0.005249743815511,
-54.21134567260742,
-0.088609740138054,
65.84819030761719,
0.32907697558403
],
"range": [
21.570192337036133,
0.547411352396012,
13.825302124023438,
1.258413732051849,
38.841251373291016,
1.565885424613953,
13.12945556640625,
0.633619353175164
]
}
And here’s my input(/testing) corpus (jongly.aif analyzed as a single pass using the same settings/hop/etc…):
{
"cols": 8,
"data_high": [
-18.309932708740234,
0.7461849451065063,
92.47969818115234,
0.6848874688148499,
-22.852352142333984,
0.35558420419692993,
116.43193817138672,
0.2502046227455139
],
"data_low": [
-29.621623992919922,
-0.7721195220947266,
47.43507385253906,
-1.7414435148239136,
-41.677947998046875,
-0.8485021591186523,
70.7255630493164,
0.045023296028375626
],
"high": 75.0,
"low": 25.0,
"median": [
-24.74309539794922,
-0.1418006867170334,
63.80019760131836,
-0.4384874403476715,
-32.07149887084961,
-0.2410660982131958,
83.63229370117188,
0.20762591063976288
],
"range": [
11.311691284179688,
1.518304467201233,
45.04462432861328,
2.4263309836387634,
18.82559585571289,
1.2040863633155823,
45.70637512207031,
0.2051813267171383
]
}
So in this case, would/could I create a “synthetic” fit by subtracting the corpus by the input, and offsetting the numbers in the input one?
So the first column of “high” in each is dB. In the corpus it’s -20.102773666381836, and the input is -18.309932708740234, so subtracting them would give me -1.792841, which would then create a new/synthetic “high” of -16.517092.
And I would repeat this for every value across the fits? (except the "high: 75 and low:25 bit)? Then load that into a fluid.robustscale~ and transformpoint jongly.aif in to “stretch” jongly to the corpus space?