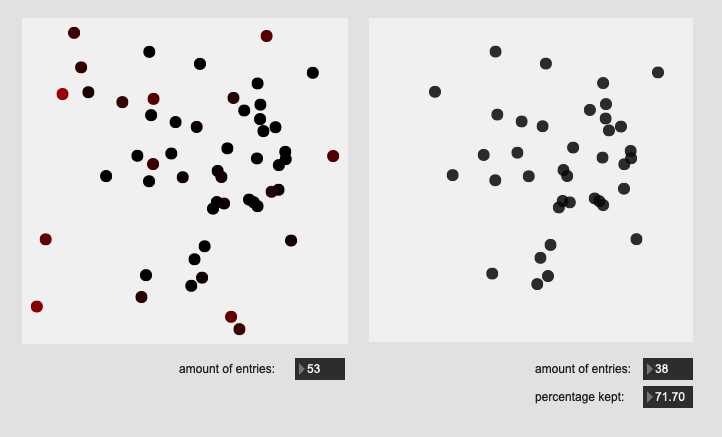
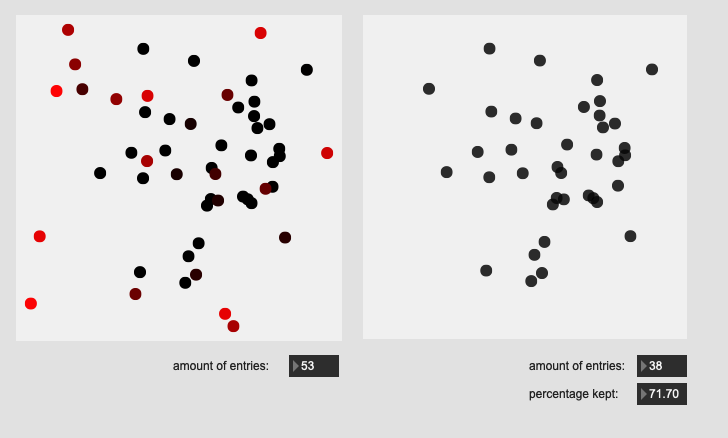
Hehe, have to leave room for future re-ification!
AMAZING!!!
This is super super super useful!
It also seems a lot more understandable than I thought it would. Don’t get me wrong, I don’t think I could connect the dots of that paper and a bunch of Max code, but walking through it all, I can follow what’s happening.
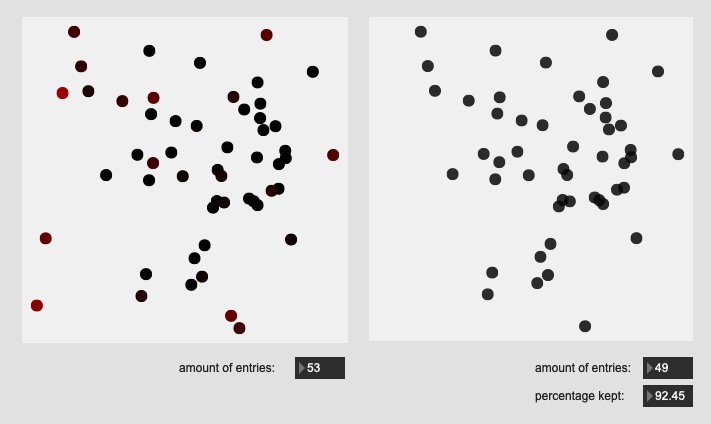
Here are some results on more real-world datasets where each class represents a bunch of actually similar sounds. The dataset above is one of these but with a single hit from a very different sounding class to act as an obvious outlier.
I’m really pleased with the (visual) results. I look forward to testing this across a few ongoing research strands.
Center hits on drum:
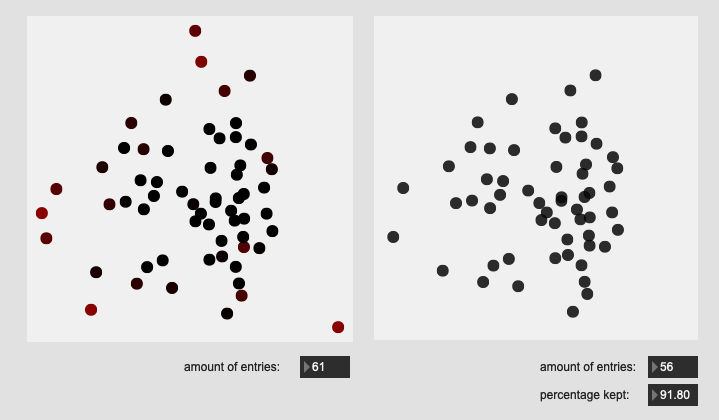
Edge hits on drum:
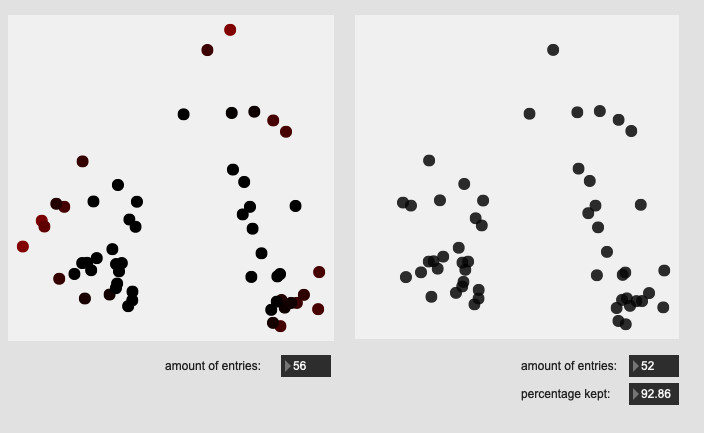
Stick shoulder hits on rim:
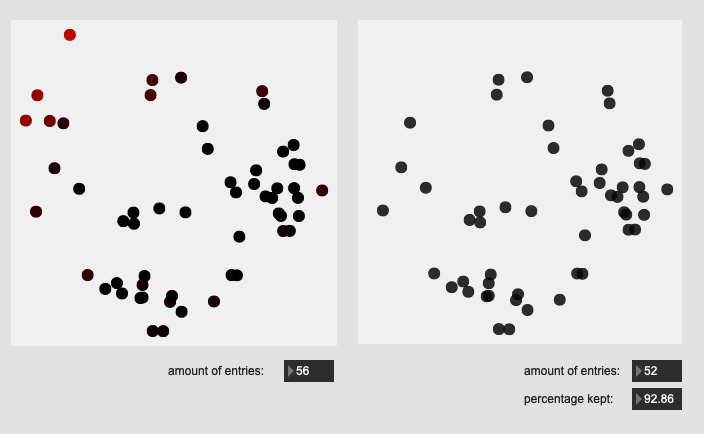
Stick tip hits on rim:
All of these are doing the outlier maths on the 104d space, then applying plotting and removing points from a PCA 2d projection.
I’ve yet to do testing to see how this effects classification accuracy, or the ability to “interpolate” between classes, but it looks like a reasonable amount of data to prune, and clustering still looks decent enough.
So a couple of follow up questions based on the patch/code.
//////////////////////////////////////////////////////////////////////////////////////////////////////////////
With regards to the numneighbours for the outlier probability computation. You say:
Size of neighbourhood for measuring local density. Has to be eye-balled really: there’s a sweet spot for every dataset. Too small, and you’ll see ‘outliers’ everywhere (because it finds lots of small neighbourhoods), too big and performance starts to degrade.
Is the only danger of “too big” being a computational hit? In looking at the code, it looks like this parameter just caps the amount of neighbors to search by, so if numneighbours > entries it does nothing. In my case I rarely go over 100 entries (per any given class) or a couple hundred in a really freak case, so would it be unreasonable just to have that value topped out at a couple hundred?
(I can imagine this mattering a lot more if you have tens of thousands of points you are trying to sort through)
//////////////////////////////////////////////////////////////////////////////////////////////////////////////
In playing with the tolerance, I’m unsure what the impact on the outliers as compared to the thresholding being applied at a fluid.datasetquery~ step.
As in, making the tolerance smaller gets more selective at the algorithm stage, with a similar thing happening at the query stage. Is there any benefit to messing with tolerance at all vs just being able to tweak the thresholding afterwards?
Here’s a comparison of the same data with different tolerance and < sttings, giving the same exact end results.
Here’s tolerance 1 + filter 0 < 0.4:
And here’s the same data with tolerance 3 + filter 0 < 0.15`:
If these are computationally the same, I’d be inclined to leave the tolerance as a fixed value, and use the more precise thresholding afforded by < to to the pruning after.
//////////////////////////////////////////////////////////////////////////////////////////////////////////////
So for the examples above, I’m applying the outlier maths to the full dimensionality, and then visualizing that in a 2d space.
Now I’m still experimenting with the accuracy of this in general, but I found I was getting slightly better classification results using PCA’d data to feed into the classifier. Similarly, I got better results for class “interpolation” on the dimensionally reduced data.
In a context where there will be data reduction happening, is it desirable (as in a “best practice”/“standard data science”) to apply this outlier rejection on “full resolution” higher-dimensional space, or the “actually being used in classification” lower-dimensional space.
I can smell an "it depends"™ a mile away here, but just wondering if there’s any established way of doing this and/or a meaningful reason to do it one way vs the other.
//////////////////////////////////////////////////////////////////////////////////////////////////////////////
Somewhat tangentially, a comment you made towards the bottom of the patch
Not implemented, but once fitted one could use this to test new incoming datapoints for outlier-ness in terms of the original filtted data
made me think of something like this potentially having implications when trying to “interpolate” between trained classes. I did a ton of testing in this other thread, but one of my original ideas was to compute the mean of each class, then use an individual points distance to that mean to indicate a kind of “position” or “overall distance”, but that ended up working very poorly.
Part of what seem to make this kind of outlier rejection work is that it’s computed across all the neighbours in a way that doesn’t get smeared/averaged like what I was getting with computing the distance of a bunch of MFCCs/stats to a synthetic mean.
If I were to take two classes worth of data and then figure out the outlier probabilities across all the points (or I guess doing it twice, once per pair?) would it be possible to compute a single value that represents how “outlier”-y a point is relative to one or the other class?
In short, is there a magic bullet to “interpolation” buried in this outlier probability stuff?
//////////////////////////////////////////////////////////////////////////////////////////////////////////////
Lastly, a fairly boring question.
Does something bad happen if you arbitrarily create gaps in a fluid.dataset~/fluid.labelset~? As in, if I use this outlier rejection stuff, and remove a bunch of entries, so I now have a dataset with non-contiguous indices, does that mess around with classification/regression/etc…
I wouldn’t think so, but then again I’ve never “poked holes” in a dataset that way.
//////////////////////////////////////////////////////////////////////////////////////////////////////////////
So the tldr of questions:
- Is there a reason to not just use
numneighbours 500all the time? - Is there a benefit of adjusting both
toleranceandfluid.datasetquery~threshold independently? - Should outlier rejection happen pre or post dimensionality reduction?
- Does an approach like this have any useful implications for computing class “interpolation”?
- Can anything bad happen in the
fluid.versewith disjointed entries in afluid.dataset~?