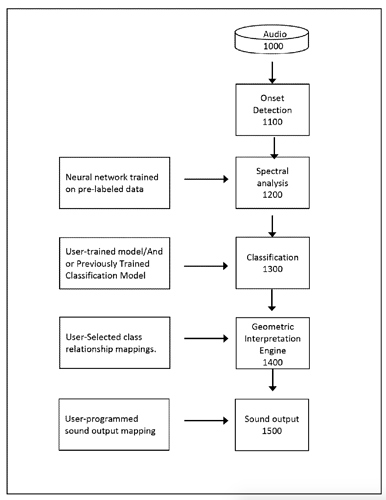
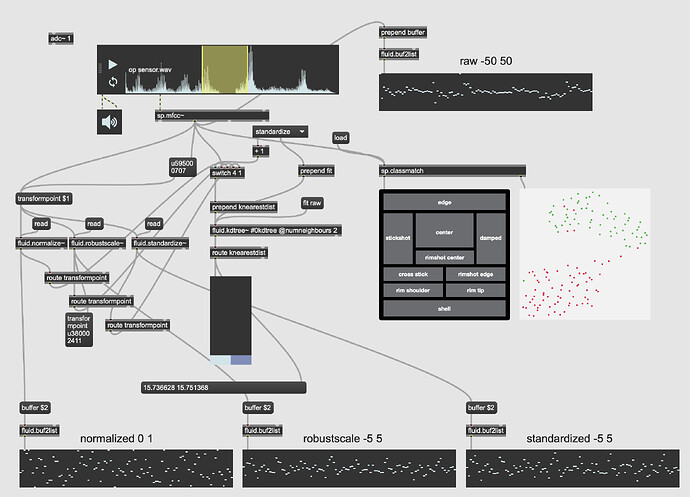
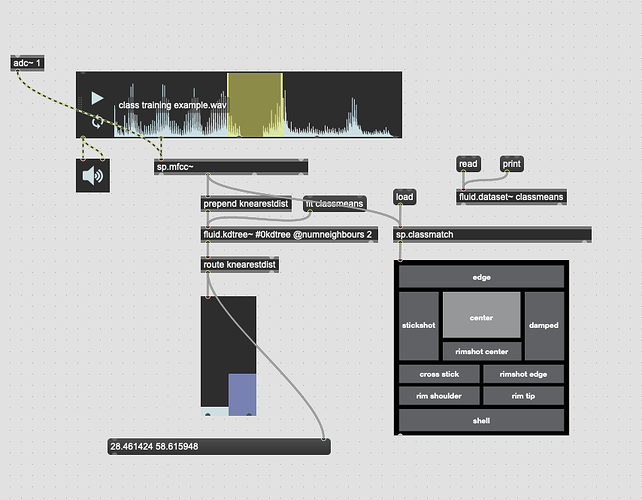
So something I’ve been trying to do for a while is be able to train specific classes and then when feeding it new audio, be able to not only retrieve the nearest class, but to also to get the distance to that class. Or rather, whether it is more one class than the other.
The first part of that is well sorted now, and I’ve even improved it recently moving over to fluid.mlpclassifier from fluid.knnclassifier, but at its core its using a 104d (12 MFCCs + stats) space to use to feed the classifier(s).
The second part of that has confused me so it’s been on the back burner until recently. After having a very useful chat with @jamesbradbury I realized the missing part of the puzzle was putting things back into a KDTree and using knearestdist to find out the distance to the points, but I’m still not entirely sure how to go about this.
/////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////
Firstly, some context.
The intended use case here is to be able to train two points like “center of the drum” and “edge of the drum” and then by knowing the distance to each class, be able to approximate the position I’m striking the drum. Similar to this:
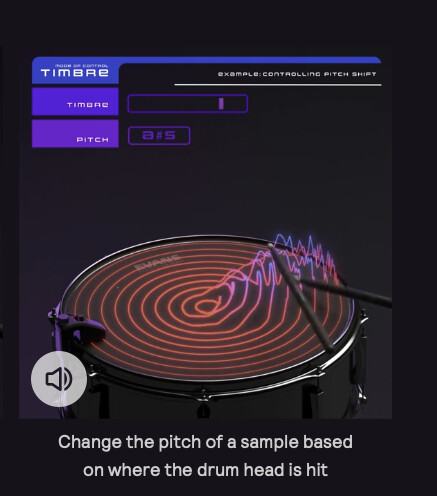
(you can hear the example about halfway down this page)
There are obviously loads more interesting applications of this, but this is my starting point as it involves just training two classes, and getting a single value that represents how close it is to one or the other.
/////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////
Now back to the problem(s)/confusion.
In looking at the fluid.kmeans~ helpfile, it looks like getmeans would be my first port of call in order to get the means of each manually labelled class but it looks like getmeans only works if you ask fluid.kmeans to create clusters for you. If you have manually created these classes, it doesn’t look like there’s a way to compute the means?
So the first problem is generating the means of each class in the first place.
Is there not a native way to do this? Would I have to dump the contents of each dataset, group them into their respective classes, then either chuck rotated versions into fluid.bufcompose~ or just doing the maths in Max-land? And for that, is it a matter of literally getting the mean of each column? (i.e. the mean of every instance of the 1st coefficient for classA would represent the mean of that point) So the end result for this process would be two entries of 104d each, with each of those ds being the mean of their respective columns?
The second problem is once the above is solved, presumably the next step is creating a KDTree with two entries, classA and classB. I would then give it new points and with a combination of knearest and knearestdist know which “class” I’m nearest, and the distance to it.
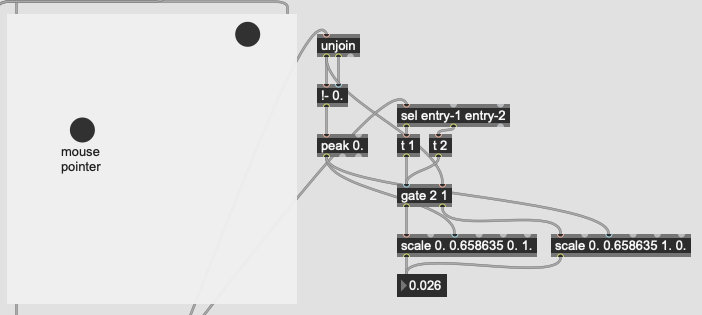
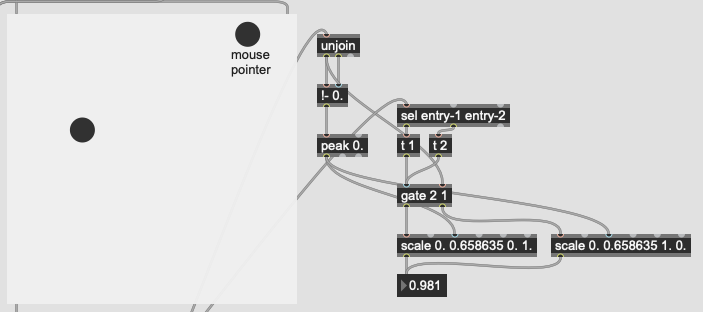
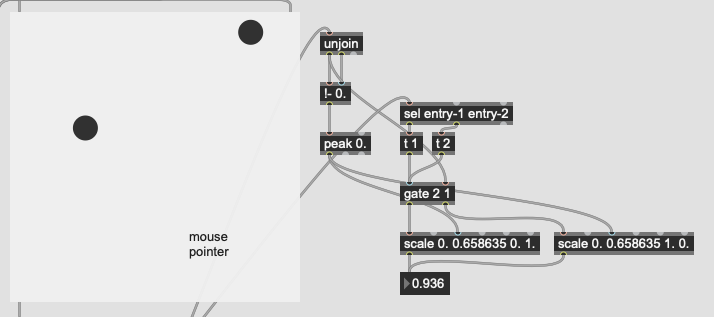
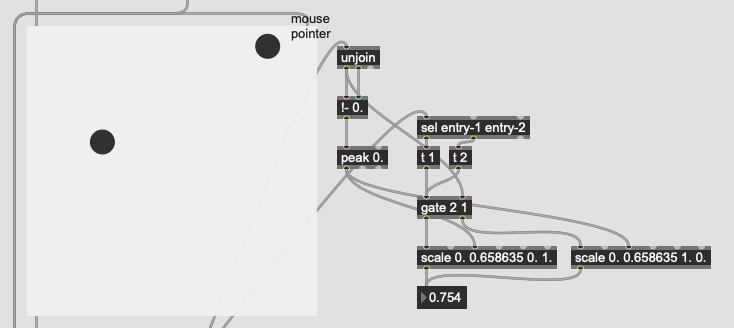
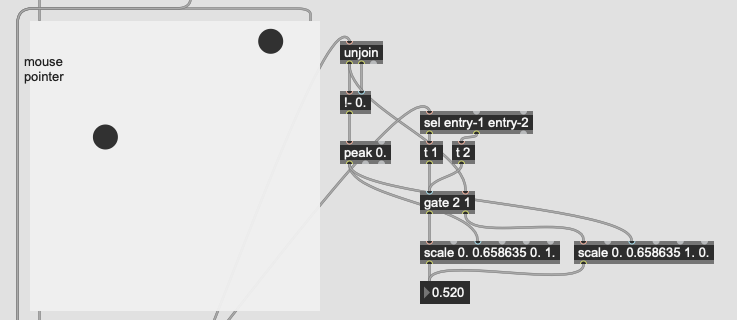
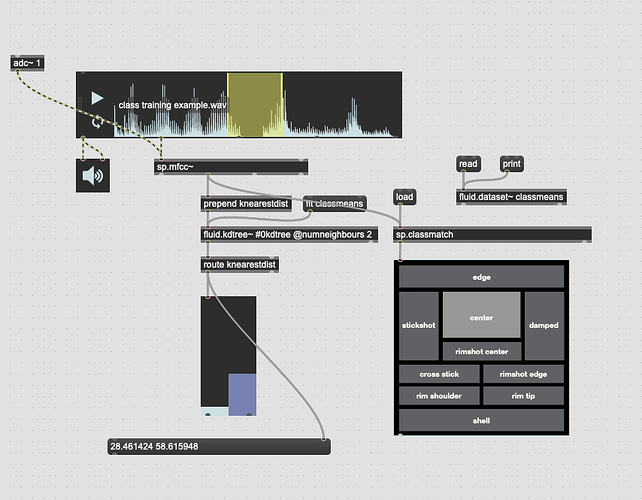
The problem I’m not getting here is that I’m not actually interested in the distance from the point I’ve matched, I want to know how much more the input is like one class or the other. So in poking around the fluid.kdtree~ help file, if I make a fake dataset with two points and set @numneighbours 2 (so I get the distance to/from both) I get something like this (with fake “mouse pointer” for the sake of legibility):
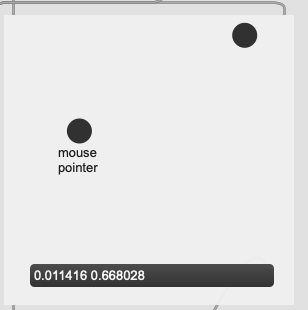

Now after a bit of logic/scaling, I can create a float where 0. = classA and 1. = classB:
I have to figure out the maximum distance between the points first, then scale accordingly.
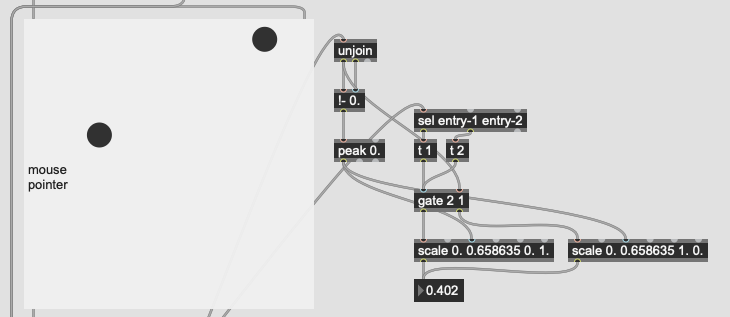
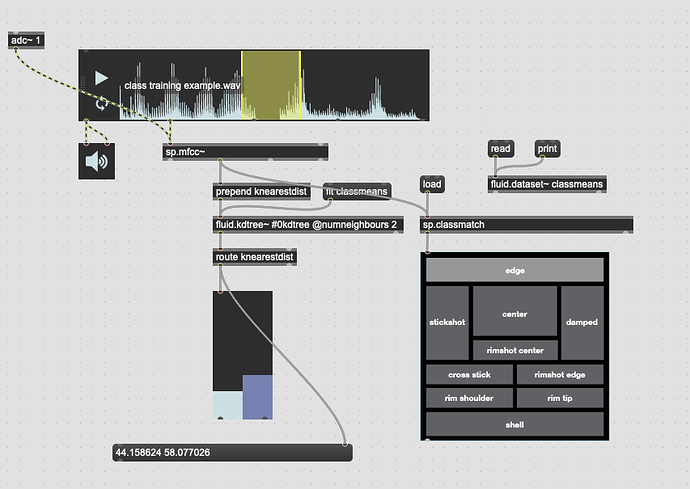
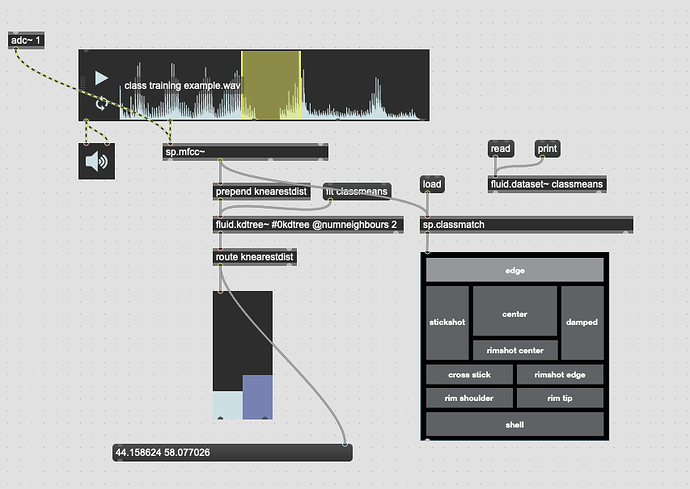
This works great… if I only move between the points. If I’m anywhere not directly on that line between them, this math/scaling doesn’t work. And also freaks out if I’m “past” the points:
There’s probably some maths here I’m overlooking to fuse/combine these numbers to get the nearest class, but in an example like this:
I would want the answer to be 100% classA (so 0. given the scaling above), since even though it is not directly on top of classA, it is much closer to it than it is to classB.
I imagine it will be something like scaling both numbers by some factor then summing them, but for the life of me I can’t think or picture what or how that would work.
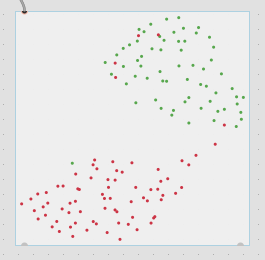
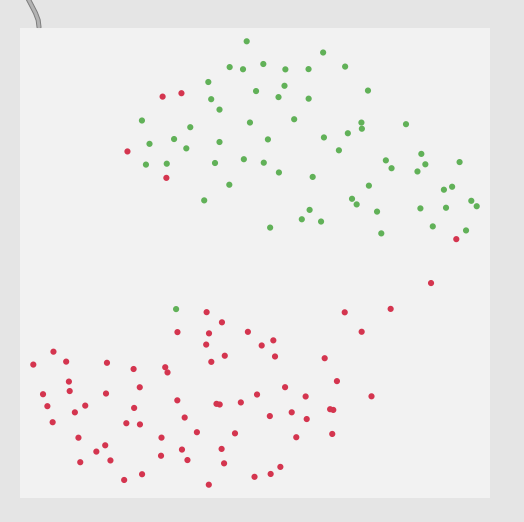
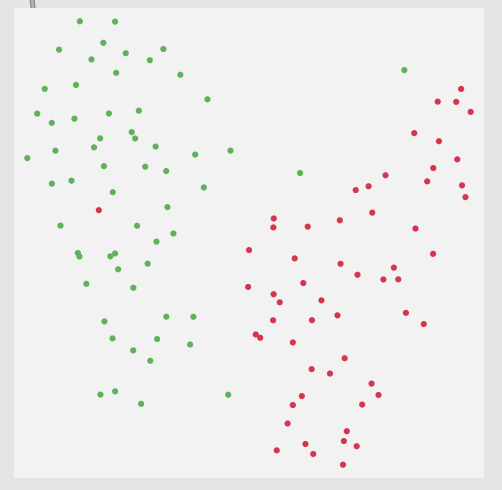
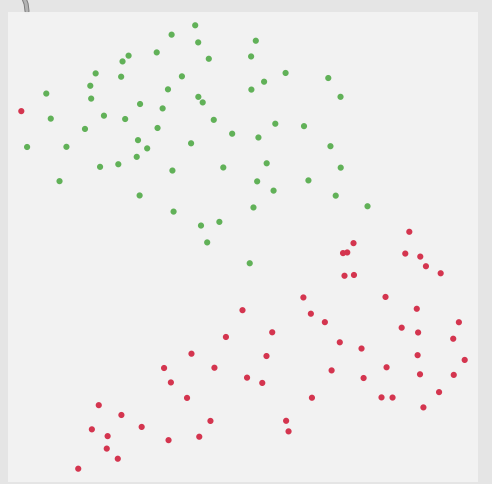
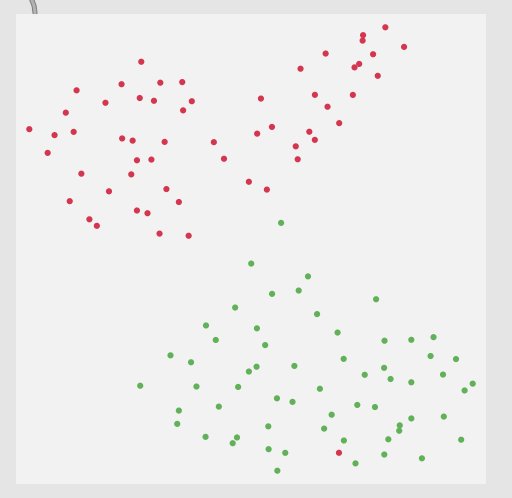
Thirdly, although this is not a full-blown problem in-and-of itself, my actual data will be 104d, so it won’t really be possible to poke/investigate it in the same way as this 2d XY example.
Here is a 2d flattening (UMAP) of the type of spread/data I’m going to be actually working with:
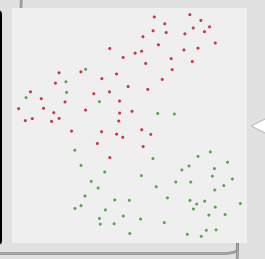
/////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////
So I guess the initial thing I’m unsure about is if I have the general workflow correct here:
- Create two classes uses
fluid.knnclassifier~with 104d input data. - Generate the means for each of these two classes…“somehow”.
- Create a two entry KDTree with the means of each class as a point.
- Get the
knearestdistof those two points and do some maths to get a float of where on the spectrum betweenclassAandclassBthe results ofknearestinputs are.
Is that about right?
If so,
- is there a sensible/native way to do step 2 (short of dumping/sorting/mathing each point in Max-land)?
- what would the maths be for step 4 that still takes into consideration the proximity to both points and returns the nearest point while taking into consideration the further one