After the tail end discussion of the previous Friday meeting I decided to implement one of the theories that @tremblap suggested for dealing with some difficult segmentation scenarios I had.
The gist of the problem that I have sounds that have areas of rapidly changing spectral material, with others being more static and immobile. I want to seperate the areas of high change from the areas of low change to give me back ‘gestures’ and ‘textures’ put crudely. I am able to find segmentation parameters (with noveltyslice~) that are quite brittle and so a small change can easily produce too many or too little segments. Under segmenting a file is a huge loss in useful information while over segmenting makes it really hard to know what is meaningful while still often giving you the right slice somewhere in the mess of things. To narrow down the error in the over segmentation which is a more ideal situation, we could cluster together contiguous slices that are too similar with a classification/clustering algorithm. That way it resolves the ticklishness of the novelty slicing while keeping the general mechanism of the novelty slicing.
I’d love some feedback or ideas on how to push this further. Some initial ideas
- Challenge the assumptions that I’m making about contiguity being important. There might be something more nuanced in looking at the distances between the slices if they are different clusters as they could be from two clusters which border each other.
- Over segmentation will generally produce contiguous chunks that are similar so its likely you’ll end up with some slices after the clustering which mean nothing.
- I’m using MFCC’s to make some analysis of the slices but there might be something else worth using
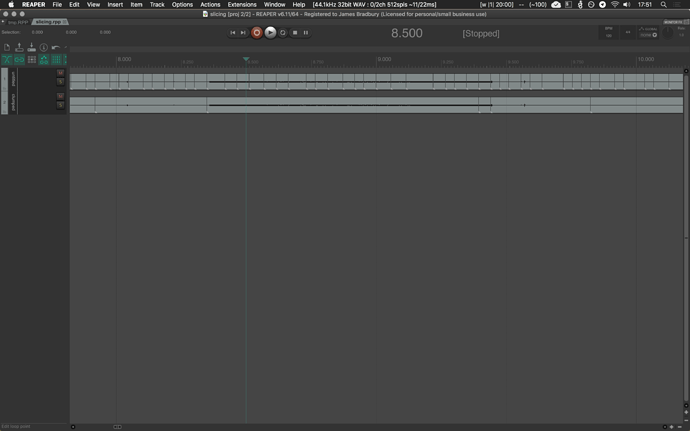
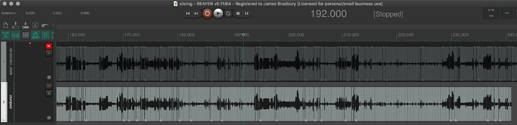
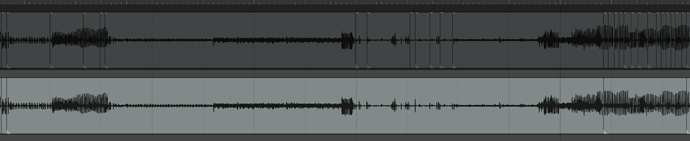
I’ll show the results in REAPER which I think speak for themselves, and then link the Python code below so anyone who wants to play with it and is so inclined that way can.
The top track is the pre-clustering
The bottom track is post-clustering
And the code is here:
import os
import subprocess
import jinja2
import numpy as np
from umap import UMAP
import umap.plot
from utils import bufspill, write_json
from slicing import segment
from pathlib import Path
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import AgglomerativeClustering
COMPONENTS = 2 # UMAP Components
NEIGHBOURS = 10 # UMAP neighbours
MINDIST = 0.05 # UMAP minimum distance
CLUSTERS = 3 # number of clusters to classify
source = Path("audio/untitled.wav").resolve()
indices = Path("/tmp/slices.wav")
mfcc = Path("/tmp/mfcc.wav")
stats = Path("/tmp/stats.wav")
output = Path("slices").resolve()
# containers for data and labels
data = []
labels = []
subprocess.call([
"fluid-noveltyslice",
"-source", str(source),
"-indices", str(indices),
"-threshold", "0.3",
"-fftsettings", "1024", "512", "1024"
])
# Analyse contiguous slice pairs
slices = bufspill(str(indices)).tolist()
slices = [int(x) for x in slices]
for i, (start, end) in enumerate(zip(slices, slices[1:])):
subprocess.call([
"fluid-mfcc",
"-maxnumcoeffs", "13",
"-source", str(source),
"-features", str(mfcc),
"-startframe", str(start),
"-numframes", str(end-start),
])
subprocess.call([
"fluid-stats",
"-source", str(mfcc),
"-stats", str(stats),
"-numderivs", "2"
])
data.append(bufspill(str(stats)).flatten().tolist())
labels.append(f"slice.{i}")
# standardise data
standardise = StandardScaler()
data = np.array(data)
data = standardise.fit_transform(data)
# dimension reduction
redux = UMAP(n_components=COMPONENTS, n_neighbors=NEIGHBOURS, min_dist=MINDIST, random_state=42)
embedding = redux.fit(data)
reduced = embedding.transform(data)
p = umap.plot.interactive(embedding, point_size=2)
umap.plot.show(p)
# umap.plot.points(mapper, labels=)
# clustering
cluster = AgglomerativeClustering(n_clusters=CLUSTERS).fit(reduced)
clumped = [] # clumped slices
cur = -2
for i, c in enumerate(cluster.labels_):
prev = cur
cur = c
if cur != prev:
clumped.append(slices[i])
# Create reaper files to look at the results
tracks = {}
pos = 0
for i, (start, end) in enumerate(zip(slices, slices[1:])):
start = (start / 44100)
end = (end / 44100)
item = {
"file": source,
"length": end - start,
"start": start,
"position": pos
}
pos += end-start
if source.stem in tracks:
tracks[source.stem].append(item)
else:
tracks[source.stem] = [item]
pos = 0
for i, (start, end) in enumerate(zip(clumped, clumped[1:])):
start = (start / 44100)
end = (end / 44100)
item = {
"file": source,
"length": end - start,
"start": start,
"position": pos
}
pos += end-start
if "clumped" in tracks:
tracks["clumped"].append(item)
else:
tracks["clumped"] = [item]
# write the reaper file out
env = jinja2.Environment(loader=jinja2.FileSystemLoader(['.']))
template = env.get_template("minimal_reaper_template.rpp_t")
with open("slicing.rpp", "w") as f:
f.write(template.render(tracks=tracks))
subprocess.call(["open", "slicing.rpp"])