As evident by some recent threads, I’ve been trying to optimize the data processing chains to avoid using dimensionality reduction in general. Among other reasons (I seemed to get worse matching overall), one of the main reasons for this was speed. Although I can’t seem to find examples/discussion on the forum as such, but I remember testing longer processing chains early on and not being very pleased with how long additional processing and dimensionality reduction processes took.
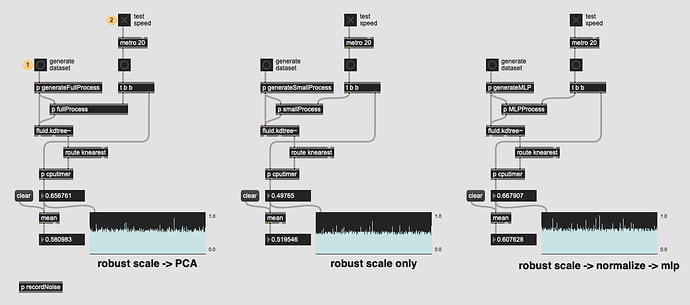
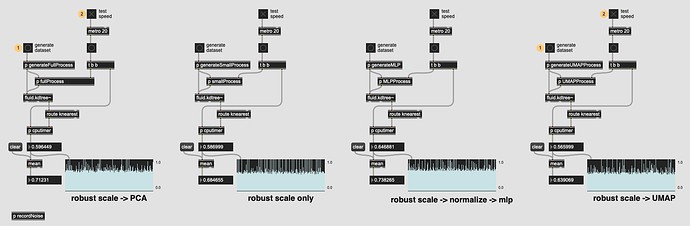
So I finally got around to building a patch that compares the raw speed of three approaches:
-
fluid.robustscale~→fluid.pca~→fluid.kdtree~ -
fluid.robustscale~→fluid.kdtree~ -
fluid.robustscale~→fluid.normalize~→fluid.mlpregressor~→fluid.kdtree~
I have to say that I’m quite pleased with the results!
The patch (attached below) creates 5000 entry datasets with 100 (randomly generated) data points and then does the associated processing/passing around.
Since the data is random the fluid.mlpregressor~ didn’t properly converge (I think I stopped training around 5.4xxxxx or so, but I figure for the purposes of speed, accuracy didn’t matter.
So it seems the PCA-ing step doesn’t add too much time at all, and MLP isn’t far behind either (though I’ve yet to get a dataset to converge using it, but that’s a different story).
Granted, in a realworld scenario there would be 10000 more steps involving individual column/stats scaling and massaging, as well as enough pruning and fluid.bufcompose~-ing to make one’s head spin, but the core speed here is pretty comparable.
I’m still a bit terrified of what *actually* processing some analysis stuff entails (a boy can dream), but for now:
speedtest.zip (144.2 KB)