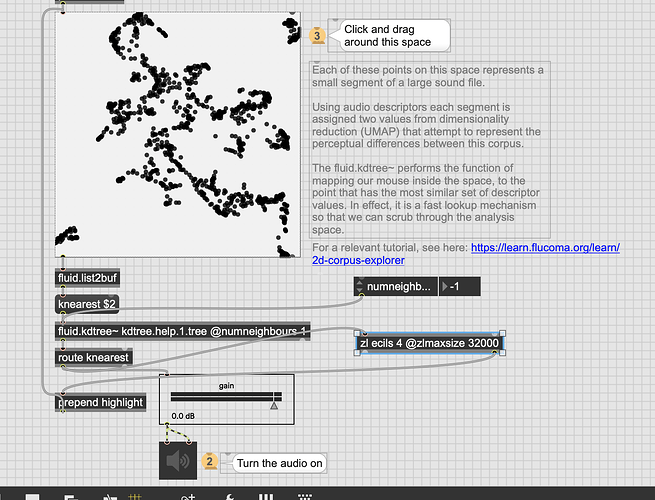
So one of the things I want to add to my dataset querying going forward is the ability to weigh some descriptors more than others.
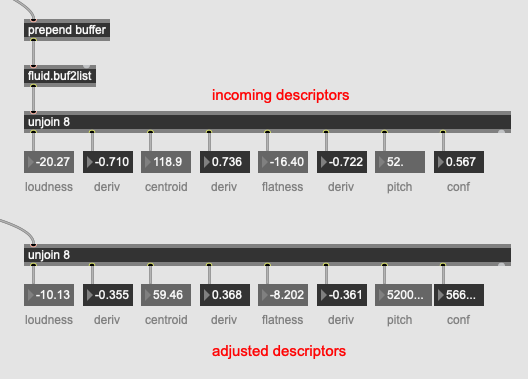
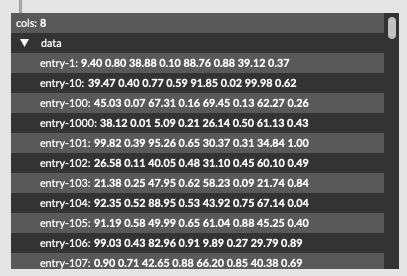
In practical terms I have 8d of descriptor data (loudness, deriv of loudness, centroid, deriv of centroid, flatness, deriv of flatness, pitch, pitch confidence), which gives me something like this for any given entry:
-23.432014465332031, 0.591225326061249, 93.79913330078125, -2.59576416015625, -12.845212936401367, -1.006386041641235, 88.705291748046875, 0.166997835040092
Loudness/flatness are in dB and centroid/pitch are in MIDI, with the derivs being whatever they feel like being.
So the core use case here would be to say something like: I want pitch to be 2x more important than other descriptors. Or I want loudness/centroid to be 10x more important than other descriptors etc…
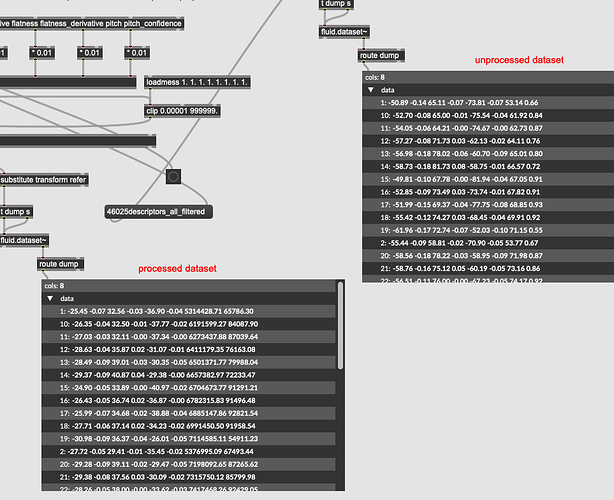
So with this, is the idea that I would scale the incoming descriptors, and then create a dupe of the dataset/kdtree with the relevant columns scaled by the same amount, then do a normal query from there?
Like if I wanted pitch to be 50% more important I would multiply my incoming pitch by 1.5x then dump/scale/re-fit the kdtree with the corresponding column also being multiplied by 1.5x?
That somewhat makes sense, though the mechanics of dumping/scaling are a bit fuzzy at the moment.
But say if I wanted to make some columns not weigh anything at all, would I zero out my input and those columns in the dataset? But if I did that wouldn’t that distance (with loads of zero values) pull a query towards them? I suppose I could just eliminate those columns, but I have logic elsewhere where I can filter by columns, so ideally I would keep the overall column/structure intact.
SO
The core questions are:
- in order to weigh a given descriptor, is it as simple as scaling up/down the incoming descriptor and corresponding dataset column by whatever amount you want?
- if that much is correct, what is the best way to scale things in this way?
I have to imagine that using one of the scaler objects (normalize/standardize/etc…) would be most efficient since dumping and iterating a dict each time I change the weighting sounds pretty rough, but I don’t want to transform the scaling overall (i.e. I don’t want to normalize or standardize my dataset). As in, I want to keep things in their natural units (dB/MIDI), so just want to scale some of those numbers. Can any of the scaler processes be fudged to do something like that? In this thread from a while back there was some good info on dumping out the scaling factors and massaging them manually, but in that case the units were actually being transformed anyways (robust scaled), so it was just a matter of adjusting that transformation. In this case I just want to apply a multiplier to some columns, straight up.