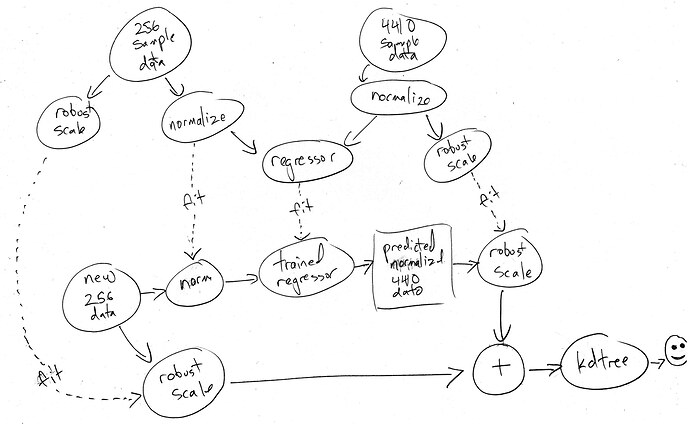
So jumping on the discussion from the LPT thread and the concept from the hybrid synthesis thread I got to thinking about how to bias a query in the newschool ML context.
For example, I want to create a multi-stage analysis similar to the LPT and hybrid approach, but a bit simpler this time. An ‘initial’ stage, and ‘the rest’. I then want to do the apples-to-orange-shaped-apples thing and match incoming onset descriptors-based analysis to find a sample from the corpus. So far so good. But what I’m thinking will be useful to do now, is have a parameter that lets me bias the querying/matching towards being more accurate in the short term vs being more accurate in the long term. Or more specifically, weighing the initial time window more, or the full sample more.
In the context of entrymatcher this would be a matter of adjusting the matching criteria by increasing/decreasing the distance for each of the associated descriptors/statistics. But with the ML stuff, it seems to me (with my limited/poor understanding at least), that the paradigm is “give me the closest n-amount of matches”, and that’s basically it. No way to bias that query (other than generic normalization/standardization stuff).
I guess I could do some of the logical database subset stuff, but that seems like it would only offer binary decision making (including or excluding either time frame).
Is there a way to do this in the newschool stuff? Or is there another solution to a similar problem/intention?