Revisiting this now in the context of fluid.normalize~ (and preset regression).
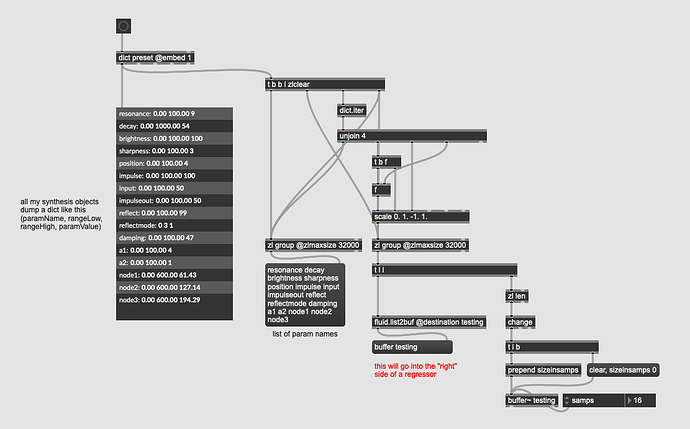
I have a bunch of canned synthesis algorithms where I’ve pre-baked the ranges, names, amount of parameters into a dict to make plugging it into a regressor fast/easy.
So far I’ve formatted things like this:
{
"resonance" : [ 0.0, 100.0, 9 ],
"decay" : [ 0.0, 1000.0, 54 ],
"brightness" : [ 0.0, 100.0, 100 ],
"sharpness" : [ 0.0, 100.0, 3 ],
"position" : [ 0.0, 100.0, 4 ],
"impulse" : [ 0.0, 100.0, 100 ],
"input" : [ 0.0, 100.0, 50 ],
"impulseout" : [ 0.0, 100.0, 50 ],
"reflect" : [ 0.0, 100.0, 99 ],
"reflectmode" : [ 0, 3, 1 ],
"damping" : [ 0.0, 100.0, 47 ],
"a1" : [ 0.0, 100.0, 4 ],
"a2" : [ 0.0, 100.0, 1 ],
"node1" : [ 0.0, 600.0, 61.428570000000001 ],
"node2" : [ 0.0, 600.0, 127.142859999999999 ],
"node3" : [ 0.0, 600.0, 194.285720999999995 ]
}
Where I have {paramName, rangeLow, rangeHigh, paramValue} with my thinking being that when I dump things out to add a fluid.dataset~ point for future regression, I can just dict.iter through that and scale accordingly:
This actually works fine, but gets a bit messier when wanting to unpack/scale the regressor output at the backside, particularly since I want it to be fast (and I don’t think dicts are fast).
Hence being reminded of this fluid.standardize~ “hack”.
Poking at the dump output of fluid.normalize~ it looks like it has a low/high value for each column, then a min/max value for the scaled output. Like this:
{
"cols" : 3,
"data_max" : [ 10329.1865234375, 21145.490234375, -2.56557559967041 ],
"data_min" : [ 463.205169677734375, 877.32891845703125, -49.015274047851562 ],
"max" : 1.0,
"min" : 0.0
}
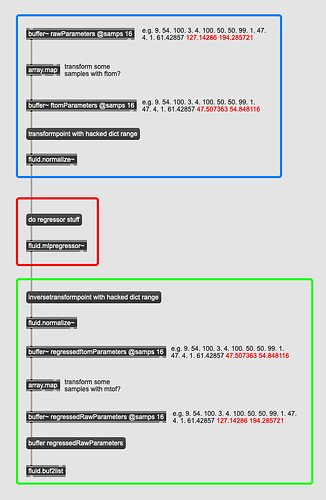
Would it just be a matter of plugging in my dict data above into this format, then transformpoint-ing the buffer~ contents on the way into the regressor then inversetransformpoint with the same fit on the way out?
That would be much simpler, and presumably faster operating on the buffer~s directy.
//////////////////////////////////////////////////////////////////
Another thing that I was thinking is that it would be ideal would be to (manually) linearize frequency-based parameters (with some ftom-ing on the way in to the regressor and mtof-ing on the way out), but I don’t know if that matters when using a regressor for synth preset morphing.
That would make things a bit stickier with using vanilla fluid.normalize~ but wondering if the new array things in Max8.6 lets you operate on buffers in this way (in a high-priority thread).
Something like this (Max pseudo code):