I know I’m quite late to the party, but based on the stuff that @tedmoore was showing in the thread about SVM stuff, I’m building a fluid.kmeans~ thing to automatically cluster a corpus to then further experiment/test on.
Other than manually making labels/classes before I’ve not done much at all with fluid.labelset~.
Now, because the visualizer I built takes a single fluid.dataset~ as its input, I want to make a 3D dataset that contains 2Ds of umap output, with the 3rd D being the normalized clusters created by fluid.kmeans~.
Seems simple enough.
But unless I’m overlooking something, do I need to have fluid.kmeans~ populate a fluid.labelset~ (with integers (as symbols)), which I then output to a dict, then uzi/iterate out the contents via a get message into a list, which I then peek~ into a buffer~, which I then have to manually add back into another fluid.dataset~ via addpoint before finally concatenating the two datasets back together. Is that right?
I understand that, fundamentally, labelsets are capable of holding symbols/names/whatever, but something like fluid.kmeans~ spits out ints anyways.
Perhaps this is just my greenness with pairing datasets/labelsets, but for the purposes of visualize at least, I kind of want all of those things in a single thing (a fluid.dataset~ in my case).
And actually, a related, but not different enough to start another thread, follow up question.
What is the intended (?) way to process/separate datasets based on information in labelsets?
More specifically:
- I have 5 sounds I want to use to create classes with my drums. I train/create the classes.
- I want to break a corpus into 5 clusters, such that each cluster corresponds with a class.
- I want to put each cluster in a different dataset (or more specifically, a different kdtree) so that I can search for the nearest neighbor within each cluster.
If I’m understanding things correctly, I need to use the labelset generated by kmeans to break the corpus/dataset into 5 separate datasets, which will each then be fit to a kdtree. So on input/analysis, I figure out what class the sound is, then once that’s determined I pass the analysis off to the relevant kdtree to find the nearest match.
So that would require me using a labelset to break apart the dataset, which leaves me in a similar predicament as above (having to data munge both the labelset and dataset).
In an ideal world the cluster/label would just be another column in the dataset that would be used for filtering(/biasing) but not distance matching. Or perhaps for distance matching with an absolute condition (cluster==5, radius==0.1, etc...). But that’s not really the paradigm.
For a small/finite amount of clusters it’s possible to create dataset forks, though this obviously gets complicated if you want an arbitrary or dynamic amount of clusters (and corresponding datasets), but that’s putting the cart ahead of the horse.
Am I simply not understanding what/how a labelset is for?
Yes, this should be easier, as it seems like a perfectly reasonable thing to do.
You can munge the dicts together, but it’s still a bit of patching. Here’s something to test against the k means helpfile:
----------begin_max5_patcher----------
2040.3oc2ZssjhaCD8YluBWj7HqGc+x7T9O1ZqoLFMLdWisKawlYSpju8HIe
YMfADCxSpZgp.eQxpOc2p6iZ4+9gEKWW9lpYYzSQeNZwh+9gEKbWxdgEcmuX
4tj2RySZbMaYg5OKW+0kqZukV8l1c4pZUkpXSTdYxl9aVreWVQtR65Hr6hYa
bs27L9DDvF0zx85iaa6kz+nR0JgKWF8ktaUknSeMqX6y0pTc6cg.DOFrJhyY
1+DD6uHbLH5K197OO7f8mU2GN+q7nl7rTUDbRThlBkzoQI5rnb00PJDQhoFj
BDVLREyARSK2sNqPEkluuQqpiJh9Ccc11sp5laA7jvCdF0AdBJlB3Tl.Qj1O
L7pHn.dW5hcplljspSTF+9MfXbPcpgR1HSMRZOIzlZcTdTt+yZgyk+Li6.oD
LGfrOB0Vk1enBli.TTowAlJwHFn8CkYLvyxj38+UVDIB3qyqTNMdwmCuqSJ1
ZMu8+mUnunJvLM0oAXtfyD9bf4dCcs5EUsulZIJryZ6hQ0kJZVv4K46y1DuI
Qmznz+q2.E9NMwuXRsqu9T4NebAr8O1bf7MYo53pjzuEYQ+SQok4MF+Ye8xE
xazVaGurxhj5e3GAjIyLgmE9HesLqvabyCqONzgHJGGif.AmJa+HLN9PJZNh
geLmjmdJ5apeLI9wSfeV3Sb0oCHX6eyjM159EkmrVk23so9LbNId3hupC3WE
7XHL1P3x3k2kGCXxigAxXDhvwvtqgL40GEEn640j7c0lmMhp4w8bh1vrb8dc
6JQVLncVrTsasxgJvptqTkTmrSYL9OqJRVmqNyM2kTUMba2cc1hOd9fbYXoP
Hc5RBBDSnDCKhNsLjOWjCu8U7v4AedVGrwsYRgbv7mPsOHCz2LqLxrlYUfow
Bh4KdTbVAW1F3gNuLopxSRUdqH.yQdFFnMFK.MGPUGs1+k.QE2nCduk9JN4B
7DwSQvXJjBkh9EKPmqY5NZU6K9IwJe0G3vFiax7JHAHVhgFu+NRURl0YfOa7
KeWJhvxvZRGBjvLk+XGh6UQb17bdSpFCC7jdXLazGCmZCMKyEOlaw7LU3cTr
.DeVRzSAwPIgPGr1j4pVAuCZlH1G.MydMg.8KDwxKE4IS6ecKfgldINlcRJH
iO3IQbnefTv.dqM3yJELlT7QSAaZEB1aEBadq1iwgoUiz6uHk++nQPdqQnyp
FQxQeztH1XFdu6D3Of.18D1IN3RGUM+eIhQWaTRpnM62U4sVGE9EDOI0PaY0
ONPMeVpI06PI.BtRX5EJPIwDrj.3cqSfJuakv4nGeK3OnaKnPLAxgb2RiIyy
Vf9pVW07ziO1jl8sL8mxUI0Ewk0aerQamq8ndutrNKI2dtNqQmklj+bVwKpZ
UQp5w8EM6qT0eOqwL020YChheUuK+25BgaN+SaM9UU1CJW2XZbhM1SymzkFd
3u9SRQ4YEpzx8E5wgtdorP+hsJEiTpVgOsLurtUqAhMNGRKwQSLI.UHbGwDH
L0Vg9QQntlwjQCo0jAcUK2vxOlP.l05zaTgV2YWtCFejzsd6AfBCfRFb0jGA
OsanaseaqS1joZ01vgm0KY44tm2y8vbYqTMQKb+Buc48nmvfj6ra8+bwtLYO
LKoBOYWLo5ayZfbUXbhFXbxGqNltUU0kUk0Ve211PCM6m1UoYo+7scpjhlgK
D+pJux6xXwmYxgP1DgmQPWkqI8DjrLFQPxG.UwNcU242lpZdYMNY3bDxr5Vi
hhOtZGnoCt6d3t3hG8xb4FL60OT60TtuNsWVGdsUh94DoMJSD7hjde3OO504
IBNzpcYapJyJzcCJDAbKHmiYlHsXtf.QTqjiEV1uX9Yt4Yr49K6DOjc6N7GA
t6ghdfBX5gxtAyijmi0RPNtUQvcU1Ey5HpfbUzD2999b5MuSQ+Hg5bFXVHzR
XOTRt2in.XPX9X6CyPA8RCRC1XccUHNHiEvGbgunOcmaKCShQbIig6iYYm4C
Mwql9l2ona2Hlqa7u7rQfDcZIuH1WpAp8UZnWT414l.gim1o27NwgSDQW0ZC
Bf0tOLnWyWNa.L2J6EfKD.ahadmRtcakupjyOzXWVuQ43.BuuoH9D6rW9leP
ZeQVF4tbh4A3d4yHPmUxL4SLb5oJFv8KtWMcXWitqQhcKAotuDk9XBXgXtnv
mbWhP3Vgg95Vc2ijOSUDgHGoODVoz4Hb.2GLhBgcykf6pz.jgPa5iWee916j
ugGiDNDXB5StNbHrS1MB8p1IbHXgh7wNgBwLY2bmqRJIDqpxVHBOn+7QEyfe
IxOBI3T5hHCYGNx7ENpnFFZuTuZ5kSJ2tz9jppuqpa5jTG9VtK4qsE3RrxcZ
VQ6ot5WrrV88r91ScWIoN80LsJUuutsdcu08JqsbWoYfK1m0EZznYMCYi9G4
GWNghjcsccWxaLaMYbRtQQ9Rx9b8gJ+CpRoqrE+7GPLc.0CEo84rBKFU80ry
0t9eFWkujzTUwgE1UXal8kCBxrGwARAfz0qVk4pNGkZSWcPygiN.ryH8Y8Z9
wNVG.XWIhtDjOPjHHLEIcE2jaq6rq3ibBEFZw5fJ1Osfc653Q0y738kanduC
UBseK3NrNuB2KtypIOZzP00sgh6xE1BxuZxiNsaCE3UZqfbaojas8SVTYiyy
D030X.NpjsX4P6OtjuGrghSa9Nb1geVw1RH5s2kylQF1NChAyX461ypMHiqf
pVgpopaiTb0c8g+4g+CvOwNlq
-----------end_max5_patcher-----------
Hot yikes! Yeah that’s “a load of stuff” right there.
I’ll have a test with this and report back
1 Like
Ok, it was not as straightforward as I initially thought either. Initially I just tried doing the example from the first post, where I just add the contents of fluid.labelset~ as an additional column in a fluid.dataset~. I managed to get all of that into a single dict, but couldn’t initially figure out how to munge them together from there. Got it in the end though.
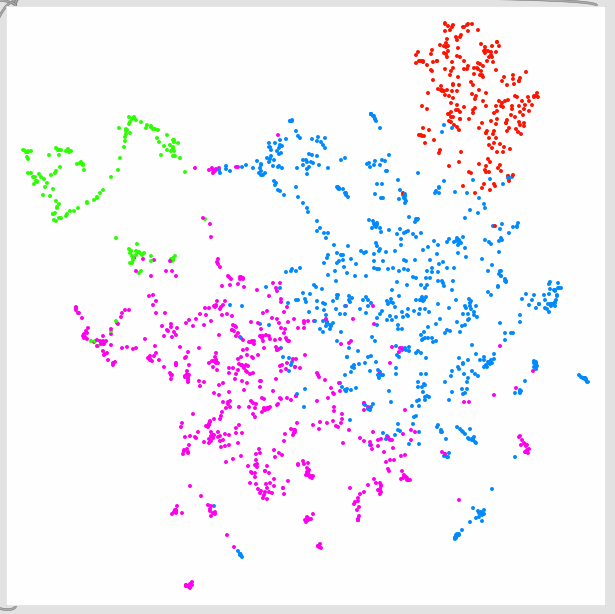
Quite handy for visualizing. This is definitely a situation where you don’t want to use the linear perception maps as the differentiation is far less with those when looking at clusters. I also noticed that if you use HSL, the edges wrap around more obviously for visualizing clusters. (I’ll update fluid.datasetplot~ in the relevant thread).
4 clusters from kmeans (linear display):
4 clusters (previous hsl):
4 clusters (new hsl):
these are sexy clusters - are you clustering after the UMAP? This is on my radar to try as it would allow to shape the space in 2-ish parameters and then get the clustering to be affected by that overall shape…
1 Like
In case I am not clear:
-
doing the clustering on the high dimension and then visualising in 2d with UMAP would give you a good way to see how the dim redux moves stuff together or apart when it was considered by KMeans in its hight dimension… that is interesting in itself.
-
doing the clustering on the reduced dimensions (post UMAP) is interesting because you can distort the reduced space to get cluster shapes and content that you like. This could be done post-autoencoder too, or post-pca or post-mds. Just fun all round.
Both interest me in my next tune, to poke at material.
1 Like
All the clusters above were done on the “raw” dimensions (21d) as I would imagine using it such a way where that is what’s in the fluid.kdtree~ as well. In other words, in this case the UMAP is just for visualization.
I haven’t played with this much at all really, but I anticipate using kmeans and such independently of any reduction, or rather avoiding reduction in general for real-time use since it’d be “slower” (the fitting of umap/etc…, but also all the pruning/peeking/composing around the process).
This sounds really interesting, but I fear by this you just mean just scaling min/max-type things (or std), as opposed to more elastically “distorting” the space, which is of definite interest.
I am talking about distortions indeed. If you check the various shapes you get in UMAP you will see you are distorting the space…
Ah right. Yeah, UMAP itself distorts the space. I thought you meant taking the projection you get from UMAP and then distorting that.
1 Like